Balancing Monoatomic Ion-Biomolecular Interactions in the Polarizable Drude ForceField
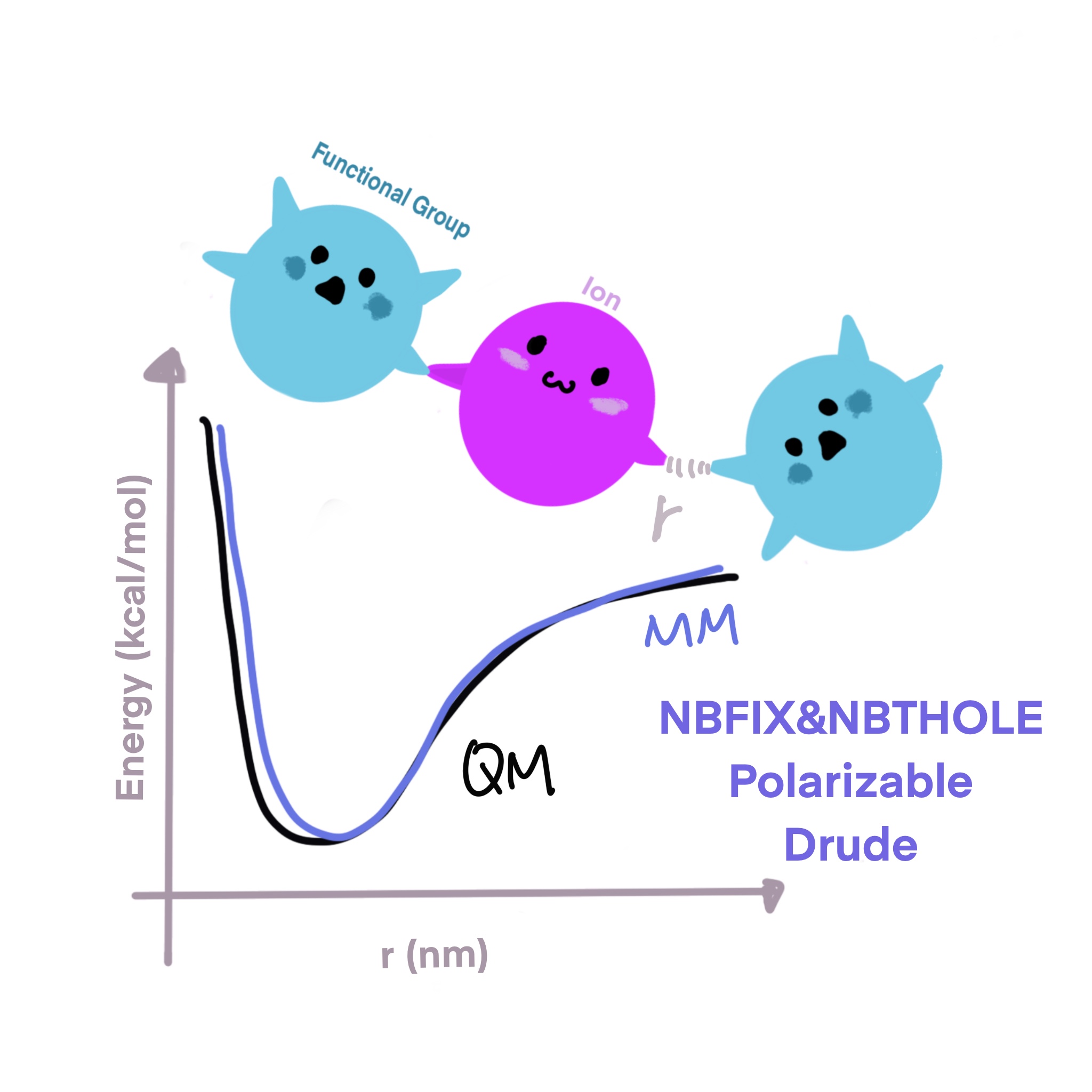
Refine the interactions between monoatomic ions (Li+, Na+, K+, Rb+, Cs+, Mg2+, Ca2+, and Zn2+) and typical functional groups present in biomolecules using a polarizable forcefield based on the classical Dude oscillator model.
University of Maryland Baltimore, Postdoc Program
- Developed software tools to automate FF parameter generation based on potential energy scans, enabling continuous FF optimization.
- Verified the proposed methodology through rigorous comparison with available experimental data (osmotic pressure, diffusion coefficient, ion competition around protein and nucleic acids etc.).
- 5 conference presentations with 1 paper published on “J. Chem. Theory Comput.” and 2 under preparation.
- Improved the FF performance in producing thermodynamic properties by up to 50 % with the new set of parameters.
- Code on working papaer is available on MacKerell Group GitHub here
